Background
Non-Targeted Analysis (NTA) is the assay of all chemicals detected by an instrument without a pre-defined list of targeted chemicals. Advancements in hardware have enabled high-resolution mass spectrometric (MS) detection, increasing confidence in molecular identifications they provide, but also causing new challenges for efficient data management and processing. Difficulty or failure occurs for ~40% of features during two-dimensional gas chromatography (GCxGC) batch NTA data processing for two major reasons. First, the mass spectral complexity results in deconvolution errors for low-level features leading to missing peaks or time-consuming manual re-integration. Second, the samples are of sufficient chromatographic complexity that data exploration results in a significant rate of divergent identifications, leaving data too complex to be reduced in a reasonable timeframe.
Approach
Our team overcomes this complexity by combining iterative processing of high-resolution GCxGC-MS data with machine learning (ML) to allow detection of low-level compounds otherwise missed by traditional peak finding algorithms. We leverage the information emergent from the batch to overcome the challenge of relying on peak finding and deconvolution for complex, high-resolution MS data. We first apply ML to automatically rank spectral signals by quality. Next, a representative signal is selected according to its quality at each chromatographic retention time in each sample. The high-resolution data is then exploited by identifying the mass spectral fingerprint of each high-quality molecular feature. This mass spectral fingerprint is leveraged in a second iteration of processing to extract quantitative information across the batch of samples by searching for specific ion signatures.
Accomplishments
Data collected from a high-resolution GCxGC-MS instrument was manually labeled according to signal quality. Both the raw and processed data were analyzed to extract predictive features. An ensemble ML model was developed, consisting of a dense neural network transferred from FloodlightTM and a convolutional neural network trained on high-resolution raw data features. Initial versions of the representative signal selection and mass spectral fingerprinting were also developed.
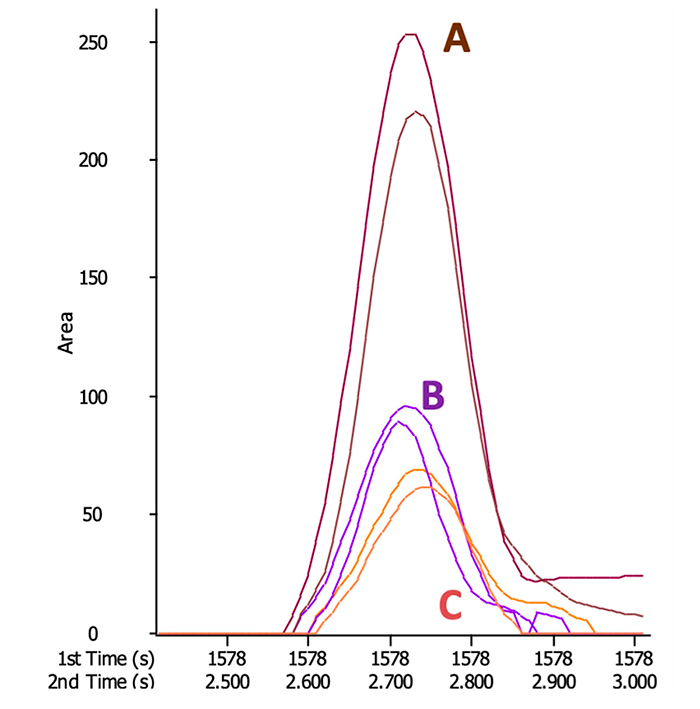
Figure 1: Low-level signal recovery (B, C) using high quality representative (A)
