Background
Trabeculae carneae are irregular muscle bundles found within the left and right ventricles of the human heart. Even though the trabecular network accounts for a significant portion of human ventricular mass, their role in the function of the ventricles is not well understood. The trabeculae carneae are difficult to resolve in clinical cardiac imaging modalities and current computational models of the heart ignore the complex trabecular structure. The goal of this project was to use SwRI’s high-resolution industrial X-ray Computerized Tomography (CT) scanner to capture detailed images of explanted human hearts. From these high-resolution CT scans, a procedure (with minimal user interaction) for characterizing the trabecular network was developed. Having these tools is the first step of larger studies to shed light on the role of the trabeculae carneae in normal and pathological heart function. Also, differences between normal and diseased hearts can be investigated, and models of the trabecular network can be included in detailed computational models of the heart.
Approach
A technique for preparing the explanted hearts for CT scanning was developed. The technique included filling the heart with expandable low-density foam that supported the interior of the heart and was invisible to X-ray energies used on this program. Explanted human hearts were scanned with an X-ray tube voltage of 120 kV resulting in images with 87-micron resolution. (Figures 1 and 2) Three-dimensional morphological image processing techniques were used to isolate the heart tissue within the image and to further isolate the internal trabecular network of the left ventricle from the surrounding heart tissue. A skeletonization algorithm was developed that simplified the trabecular structure into a series of single-voxel paths with identified branch points. (Figure 3) The skeletonization allowed for measurements of properties such as number of trabeculae, and the length and central cross-sectional area of each trabecula.
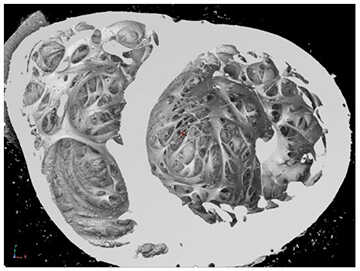
Figure 1: Surface rendering of the inside of the heart. This is a view looking toward the apex of the heart with the left ventricle on the right side of the image.
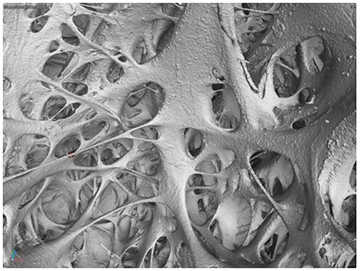
Figure 2: Visualization of inside the left ventricle near the apex.

Figure 3: Visualization of calculated skeleton (light traces) within a subset of trabecular volume.
Accomplishments
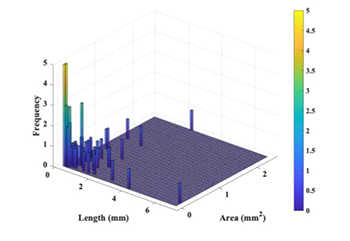
Figure 4: Length and cross-sectional area distribution of the trabeculae carneae at the apex part of the left ventricle.
A procedure for high-resolution ex-vivo imaging of internal heart structures was developed. CT images were acquired from six human cadaver hearts. Of those hearts, three were from people who suffered from heart disease and three did not. Statistics of trabecular length and width for one of the six hearts was performed. (Figure 4) Follow-on study of the remaining hearts and future application of the developed algorithm will be part of the Ph.D. research for at least one UTSA graduate student. Results of this study will be carried forward into future projects where the trabecular structure will be included in computerized modeling of heart function.
