Background
Despite substantial advancement in genetic discoveries, our understanding of environmental risk factors and the exposome remains limited. With overarching differences in individuals and their environments, the chemical composition of the environment coupled with metabolomic information can lead to discovery in the field of personalized medicine. Findings suggest human hair may be promising biospecimens for monitoring long- and short-term exposure to multiple environmental chemicals and perturbations in endogenous chemicals in biomonitoring investigations. Hair is an attractive matrix for the proposed work because it collects chemicals both from the internal, biological source and from external exposures through contact with vapors and liquids.
Non-targeted analysis (NTA) and suspect screening analysis (SSA) are two powerful techniques relying on high-resolution mass spectrometry (HRMS) and computational tools to detect and to identify unknown or suspected chemicals in the exposome. SwRI's Lighthouse™ software suite utilizes machine learning algorithms to efficiently process SSA and NTA methods. This technology was used to extract chemical exposure data from hair samples for metabolite fingerprinting.
Approach
The proposed research tested two hypotheses: 1) health outcomes among individuals are partially attributable to differences in chemical exposure, and 2) environmental and metabolic biomarkers of differences in chemical exposure are accessible through hair samples. Our approach would attest to the feasibility of using hair samples to predict health outcomes more accurately among individuals by attributing their chemical exposure history, allowing us to propose a new diagnostic assay and investigation of individual variation in exposure fingerprints and health outcomes.
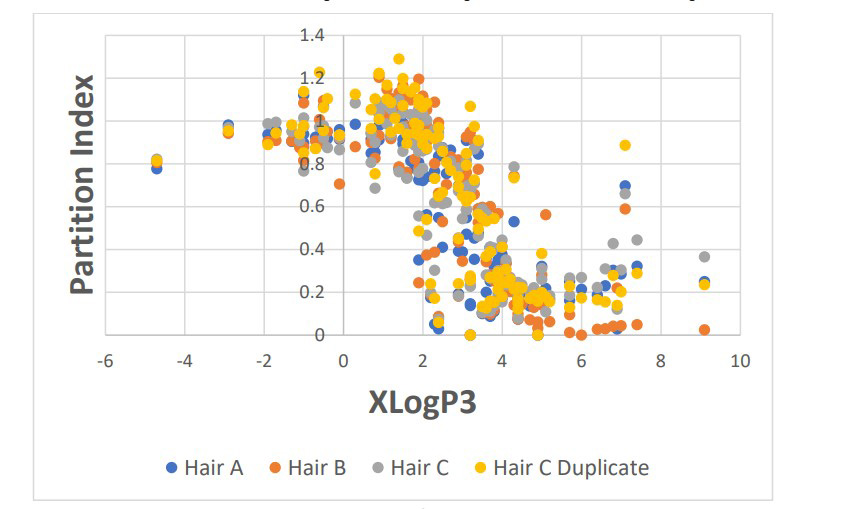
Figure 1: Partition index versus XLogP3.
Accomplishments
We surpassed our classification metric targets: accuracy of 90%, F1-score, and area under the receiver operating characteristic (AUC/ROC) of 85%, achieving an average accuracy of 93%, F1-score of 94%, and ROC-AUC of 100% overall health outcomes in the test set for Maryland (MD). Our models over the Arizona (AZ) data performed worse due to less data availability, with an accuracy of 70%, F1-score of 47%, and ROC-AUC of 60%. We saw similar results for the regression models, having a Mean Squared Error (MSE) of 1.47 for MD and 2.35 for AZ. Both regression model predictions had high correlation with ground-truth labels.
For the chemical analysis portion of this research, the interaction with hair from an aqueous solution was studied for 124 chemicals using three different analytical methods as described above. The octanol/water partition coefficient (XLogP3) as calculated by PubChem ranged from -4.7 to +9.1. The partition index as a function of XLogP3 is shown in Figure 1, with any native signals detected in the control hair sample subtracted from the spiked hair. An inflection point is observed around an XLogP3 value of 3.0 indicating a stronger affinity for hair. Overall, 89 of the 124 chemicals studied (72%) were recovered from the hair. This can be used predictively to model which chemicals can be monitored in hair as an indication of environmental exposure.

