Background
Cancer is one of the leading causes of death worldwide, accounting for 10 million deaths in 2020 alone. The most common type of oncology therapeutics are DNA-targeting small molecules, which either damage DNA or inhibit its replication (Figure 1). Since the growth rate of cancer cells are often much higher than healthy cells, and often have compromised DNA repair machinery, these cells are much more sensitive to the damaging effects of these therapeutics. Unfortunately, DNA-targeting drugs are notorious for side effects and have even been known to induce malignancy themselves. Therefore, the design, testing, and development of more selective, safer DNA-targeting cancer drugs remains at the forefront of pharmaceutical research and development. Yet, compared to the computer aided design of protein-targeting small molecules, the field of DNA-targeting small molecules has been relatively undeveloped. To address this unmet need, SwRI recently completed an internal research and development program to utilize our machine learning (ML) guided docking program, Rhodium™, to predict the binding affinity and cancer cell cytotoxicity of known DNA-targeting small molecules.
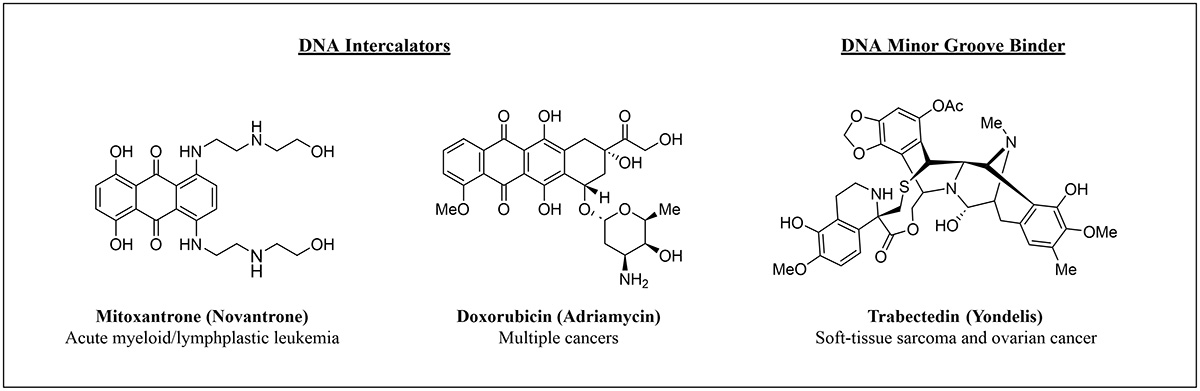
Figure 1: FDA-approved DNA targeting oncology drugs.
Approach
Traditionally, Rhodium™ and other reported docking programs have only been statistically validated for studying protein-ligand interactions. Thus, before this work there were no clear examples of a validated ML-guided docking platform for structure-based drug design of ligands that target DNA. To train Rhodium™, a virtual library of highly trustworthy DNA crystal structures was established as well as unbiased small molecule datasets. This database was segregated into several training sets, each serving unique roles in screening potential DNA-targeting oncology drugs, such as predicting the binding geometry of DNA interactions for both intercalating and minor groove binding ligands (Figure 2), binding affinity, and anti-cancer activity.
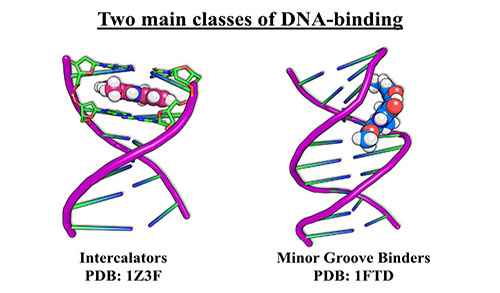
Figure 2: Two most common binding modes of small molecules with DNA.
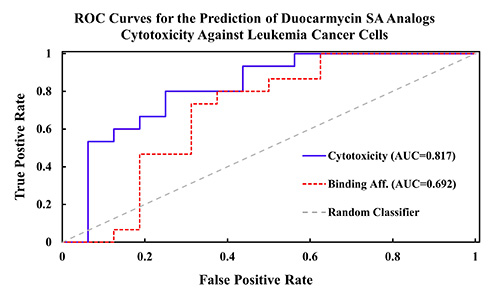
Figure 3: ROC curves demonstrating the enhanced true positive rate vs false positive rate when using our developed training sets to predict the rank ordered activities of a series of 31 duocarmycin SA analogs against leukemia cancer cells (L1210).
Accomplishments
SwRI demonstrated the efficacy of this technology by rank ordering the potency of compounds that exhibit anti-cancer activity in several cancer cell models, including leukemia, breast cancer, and liver cancer. A noteworthy example is the predicted rank ordering of a series of duocarmycin SA analogs, a series of picomolar DNA inhibitors which have advanced to Phase II clinical trials for cancer treatment. Using Rhodium™ and a training dataset of duocarmycin derivatives, we were successful in a validation experiment in ranking potent derivatives over inactive ones in a series of 31 test compounds with a confidence level of 99.95% (Figure 3). This promising result suggests Rhodium™ is capable of correlating DNA docking data to anticancer potency. Thus, in tandem with our validated protein docking methods, we have developed a strong platform for the discovery of future DNA-targeting cancer therapeutics.

